A grappling hook for bacterial motility uncovered
Scientist at the California Institute of Technology (CalTech) and the Max Planck Institute for Terrestrial Microbiology in Marburg Germany have determined the architecture of large protein complex that bacteria use for type IV pilus dependent motility, virulence and biofilm formation. The work can potentially lead to the development of novel treatments of bacterial infections.
Most bacteria are able to actively move from one location to another and bacterial motility is important for virulence and biofilm formation. So, the molecular machines that drive motility have become attractive targets for novel antibacterial drugs. Intriguingly, different bacteria have different machines for motility. The two best studied of these machines are flagella and type IV pili. While scientists have a very good understanding of the structure and function of the machine that drives rotation of flagella to push bacteria forward, the way the type IV pilus machine works to pull cell forward is not well-understood. Scientists at CalTech and the Max Plank Institute for Terrestrial Microbiology have now used high resolution cryo–electron tomography to determine the architecture and overall structure of the type IV pilus machine in the bacterium Myxococcus xanthus. The work, which lays the foundation for the development of novel antibacterial drugs, is published in Science.
“Type IV pili are highly dynamic filaments that are extended from the cell, attach to a surface, and then they retract. Because they adhere strongly to the surface, a cell is pulled forward during retractions. Myxococcus cells have 5-10 pili and as these pili extend and retract, the bacterium moves across a surface like a small Spiderman” explains Lotte Søgaard-Andersen, one of the leading authors on the paper.
The molecular machine that drives the extension/retraction cycle of type IV pili in Myxococcus as well as in other bacteria consists of 10 different proteins that assemble to form the machine. Recently, the Søgaard-Andersen lab was able to decipher the order in which these proteins become incorporated into the machine.
However, the actual structure of the machine still remained mysterious. The laboratories of Grant Jensen at CalTech and Lotte Søgaard-Andersen as well as colleagues at University of Utah have now solved this mystery using cryo–electron tomography.
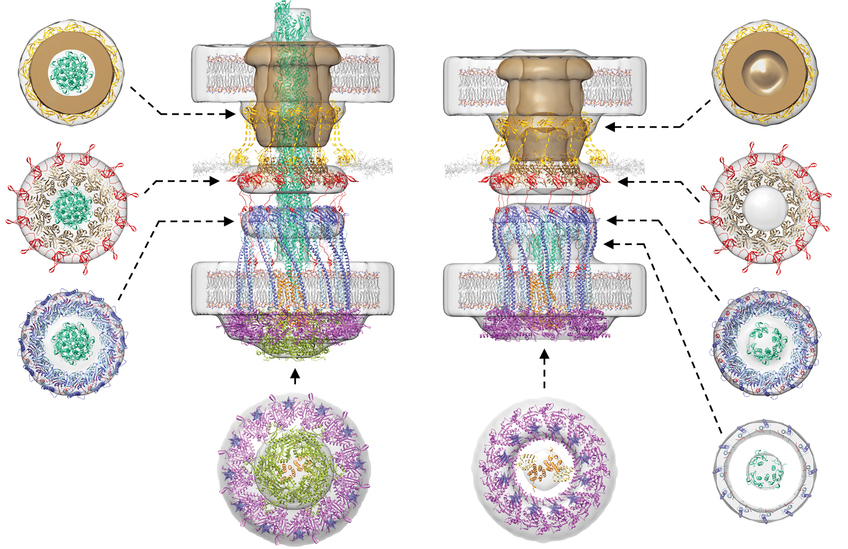
The cryo–electron tomography technique allows to record high resolution images of intact frozen cells. When this technique was applied to Myxococcus cells in the lab of Grant Jensen at CalTech, the second lead author on the paper, the filamentous type IV pili and the machine at the base of the pilus were immediately visible. “The images clearly showed that the intact machine spans the entire cell envelope” says Grant Jensen. Next, the scientists imaged mutants that either lacked a part of the machine or contained individual components fused to a fluorescent protein. By comparing the images of the intact machine with those of the modified machines, the locations of all 10 proteins in the machine were revealed at a resolution of 2-5 nm and giving the first ever glimpse of the overall architecture of the type IV pilus machine.
Wanting to get a full picture of how the machine works, the team followed up with a tour-de-force in molecular modeling. “Because we know a lot about the atomic structure of the 10 proteins that make up the type IV pilus machine, we were able to take these known structures and fit them into our architectural model to generate “pseudo-atomic” working models of a machine with an extended pilus and an empty machine without a pilus” explains Grant Jensen.
The work provided unique insights into the mechanism of the type IV pilus machine. Specifically, it appears that through ATP hydrolysis, the ATPase PilB in the cytoplasm rotates the inner membrane protein PilC. PilC functions like a paddle and scoops a PilA pilus subunit out of the inner membrane and onto the base of the growing pilus during each step in the rotation. Conversely, pilus retraction is driven by the cytoplasmic PilT ATPase that rotates PilC in the opposite direction in that way removing PilA subunits from the base of a pilus and back into the membrane.
Understanding how the type IV pili machine works can help scientists design more efficient antibacterial drugs. “Combating bacterial infections in the future may not only rely upon interfering with essential processes to kill them but also on interfering with processes to make them less fit” says Grant Jensen and Lotte Søgaard-Andersen “So we need to understand these processes – and this work provides a major guideline for how to interfere with type IV pilus function”.
This work represents a collaboration of CalTech’s Division of Biology & Biological Engineering, the Max Plank Institute for Terrestrial Microbiology in Marburg, Germany and the University of Utah in Salt Lake City, USA. The study was funded supported by the NIH, the Howard Hughes Medical Institute, the Max Planck Society, and the Deutsche Forschungsgemeinschaft within the framework of the Collaborative Research Center (SFB) 987 “Microbial Diversity in Environmental Signal Response”.
Reference
Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Søgaard-Andersen L, Jensen GJ. Architecture of the type IVa pilus machine. Science 351: aad2001 (2016). DOI: 10.1126/science.aad2001.